Prof. Wen-Jun Li's group at School of Life Sciences isolated microorganisms from hot spring microbial mats with a “combination” strategy: a perspective of microbial interaction
Source: School of Life Sciences
Written by: School of Life Sciences
Edited by: Tan Rongyu, Wang Dongmei
Microbes are creatures with the widest living range and the most abundant biological diversity on the earth. They are important material basis for human survival and important source for biotechnology innovation. However, due to the limited knowledge on the majority of microbial species, the “Great Plate Count Anomaly” is still a puzzle up to now, and hitherto uncultured microorganisms are vividly imaged as microbial “dark matters”. We do, however, understand that these “dark matters” play unprecedented roles in carbon and nitrogen cycling, novel natural products chemistry and in maintaining the balance of the environment. To culture and effectively describe the vast majority of microorganisms has always been a challenging subject in microbiology research, as biologists often say, “To really know them, you have to grow them”.
In the new study, to cultivate previously uncultured Chloroflexi microorganisms, total 26 hot spring microbial mat (HSMM) samples were collected from Yunnan and Tibet hot springs, and a co-occurrence network analysis was conducted based on the 16S rRNA genes high-throughput sequencing. Results show peripheral nodes in the network comprised of genera Chloroflexus (13.9%), Thermus (11.05%) and Roseiflexus (8.44%), and members with low abundance is the key nodes. Among these key nodes, genera Tepidimonas, Geobacillus, Meiothermus and Sphingomonas have been previously isolated. After screening tests, strain Tepidimonas SYSU G00190W (190W) was confirmed as a growth promoting strain. Spent culture medium (SCM) based on 190W was prepared and members of the phylum Chloroflexi were frequently isolated on the SCM plate, novel isolates related to Roseiflexus spp. and Chloroflexus were isolated only on the SCM plates, most of these isolates represents the novel species in the phylum Chloroflexi, two of them are distinct lineages in the phylum Chloroflexi.
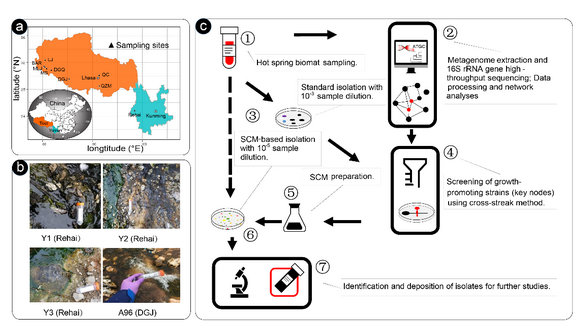
Fig. 1. Overview of the sampling sites and the workflow for the network-directed isolation procedure from HSMMs. (a) Location of the sampling sites; (b) Representative samples (Y1, Y2 and Y3 from Yunnan, and A96 from Tibet hot springs) that are used for isolation; (c) Workflow use to predict key-node taxa and screening for growth-promoting strains. SCM plate was effective in the isolation of targeted microorganisms from the HSMMs.
As the growth-promotion on Chloroflexi spp. was observed with SCM, we hypothesized that Tepidimonas sp. may have a strong ability to synthesize and excrete compounds that facilitate the Chloroflexi bacteria. Subsequently, non-targeted extracellular metabolomes was conducted on 190W and 83 low molecular weight organic substances (LMWOS) were detected to significantly accumulated during its growth. Representative LMWOS was further tested on the newly isolated strains, result showed that pantothenic acid (VB5) and 3-indoleacetic acid (IAA) facilitate growth of all the tested strains. Imidazole acetic acid and deoxycytidine showed enhance growth to Chloroflexus strains while putrescine improved the growth of Roseoflexus strains. Further, uracil, cytosine and adenine also facilitate the growth of other Chloroflexi.
This work provides strong experimental evidence that key node bacteria is critical for rapid and easy cultivation of hard and slow-growing strains from HSMMs in the laboratory condition. The study further provides an idea for cultivating of unculturable microorganisms in other environments.
Detailed information could be found at DOI: https://doi.org/10.1038/s41522-020-0131-4
The research team of Prof. Wen-Jun Li at School of Life Sciences of Sun Yat-sen University have been focusing on the extremophiles and their ecology for more than twenty years. Details of his previous researches could be found with following information.
1. Insights into ecological roles and evolution of methyl coenzyme M reductase containing hot spring Archaea.
https://www.nature.com/articles/s41467-019-12574-y
2. Genomic inference of the metabolism and evolution of the archaeal phylum Aigarchaeota.
https://www.nature.com/articles/s41467-018-05284-4
3. Discovery of Druggability-Improved Analogues by Investigation of LL-D49194α1 Biosynthetic Pathway.
https://pubs.acs.org/doi/abs/10.1021/acs.orglett.9b00610
4. Discovery and Biosynthesis of Atrovimycin, an Antitubercular and Antifungal Cyclodepsipeptide Featu-ring Vicinal-dihydroxylated Cinnamic Acyl Chain.
https://pubs.acs.org/doi/abs/10.1021/acs.orglett.9b00618
5. Update on the classification of higher ranks in the phylum Actinobacteria.
https://www.ncbi.nlm.nih.gov/pubmed/31808738
Link to the Prof. Wen-Jun Li's Lab: http://liactlab.sysu.edu.cn/index-en.html



